How Much New Knowledge is Hidden in Scientific Text?
Given the rapid pace of progress within natural language processing, it seems like the automation of scientific hypothesis generation could be feasible. But if this were possible, would we ever know?
Imagine that the pieces of a puzzle are independently designed and created, and that, when retrieved and assembled, they then reveal a pattern-undesigned, unintended, and never before seen, yet a pattern that commands interest and invites interpretation. So it is, I claim, that independently created pieces of knowledge can harbor an unseen, un-known, and unintended pattern. And so it is that the world of recorded knowledge can yield genuinely new discoveries.
Don R. Swanson
One model of a scientist is as a person that reads a lot. If one accepts the scientific method, as displayed in the figure below, as an accurate portrayal of a scientist’s job, then the steps “Observation/question” and “Research topic area” imply a lot of reading of other scientists’ “Report conclusions” steps before a “Hypothesis” can be formed.

If we assume science to be a completely closed system in which our model scientists are confined to windowless labs—their knowledge about the outside world derived only from a library of scientific papers—then the new observations made by these scientists would necessarily be dependent on the reported conclusions of other scientists.
This is not a terrible agent-based model of science. Indeed, real-life scientists do predominantly report scientific conclusions in writing, disseminating these insights in natural language through journal publications and conference proceedings. Many real-life scientists spend a nauseating amount of time confined in windowless rooms, and many enjoy passing the time by browsing these publications. Scientists are predominantly rewarded based on the quantity and quality of their written conclusions, so if we endow our model scientists with a similar publication objective, this economy of model scientists contains all of the elements necessary to reflect the cyclic publishing dynamics we observe in reality.
But it’s also not a perfect model. Scientists do, in fact, spend time outside of their windowless labs. They can make observations about the world which do not come from something they’ve read. They have friends and colleagues with whom they sometimes discuss these observations, and these discussions are usually not written down or published. In fact, if you talk to a more senior scientist, they are likely to tell you that they rarely have the time or desire to read papers at all (Hubbard & Dunbar, 2017), and instead tend to derive and communicate observations by email or through osmosis with students and collaborators. Some scientists opt out of the public scientific ecosystem altogether, avoiding the final step in the scientific method for reasons of financial gain or national security.
Despite these shortcomings, the simplicity of this agent-based model of science has appealing computational properties. Some combinatorial theories of innovation suggest that novel discoveries result from unique combinations of pre-existing concepts (Weitzman, 1998; Uzzi, Mukherjee, Stringer, & Jones, 2013), so we may not even need to assume these scientists are competent; we just need enough of them to efficiently explore the combinatorial space of plausible hypotheses. Given the recent growth of natural language processing capabilities, this simplified model’s sole reliance on scientific text as the basis for new scientific observations prompts a question: could we automate this model of scientists and extract novel scientific observations from the scientific literature alone?
Don Swanson, Arrowsmith, and early literature-based discovery
One of the first people to investigate the potential for automating scientific discovery was Don R. Swanson. An early computational linguist and information scientist, Swanson laid the foundations of what would become the field of literature-based discovery in a 1986 paper titled “Undiscovered Public Knowledge” (Swanson, 1986). In the paper, Swanson argues that the distributed and depth-first organization of the scientific enterprise generates significant latent knowledge which, if properly retrieved and combined, could lead to new scientific discoveries.
After laying this philosophical groundwork, Swanson transitioned to proving, through a series of articles in the late 1980’s, that this undiscovered public knowledge does indeed exist. Embodying our model scientist trapped in the lab with only scientific literature to occupy the time, Swanson postulated that the existence of articles linking two concepts A ↔ B and another collection linking B ↔ C indicates novel hypotheses may be found by investigating the yet-undiscovered link A ↔ C. His early implementations of this idea were successful. Noting that the literature on fish oil described outcomes related to improved blood circulation and that the literature on Raynaud’s syndrome described a blood circulation disorder, Swanson hypothesized in 1986 that fish oil could be consumed to ameliorate the symptoms of Raynaud’s syndrome (Swanson 1986). Noting that magnesium produces physiological changes related to those associated to migraine susceptibility, Swanson proposed in 1988 that magnesium had potential as a supplement for alleviating migraines (Swanson, 1988). In 2006, Swanson proposed that endurance athletes could be at increased risk of suffering atrial fibrillation, hypothesizing inflammation as the mediating link (Swanson, 2006). Each of these literature-driven hypotheses were later validated in subsequent clinical trials (DiGiacomo, Kremer, & Shah, 1989; Gordon & Lindsay, 1996; Gallai et al., 1992; Mont, Elosua, & Brugada, 2009).
Swanson continued publishing hypotheses according to his “Swanson linking” procedure (Swanson, 1987; Swanson, 1989; Swanson, 1990), eventually culminating in the creation of Arrowsmith (Swanson & Smalheiser, 1997; Smalheiser et al., 2006), a computational implementation of Swanson’s approach to literature discovery which, given text-based topics A and C, searches for topics (B) that co-occur with both A and C throughout the literature. This approach to literature-based discovery is now better known as the ABC procedure.
While Swanson’s pioneering program for generating hypotheses from pre-existing academic literature highlighted a number of new medical relationships, this initial success was highly dependent on the choice of A and C concepts to serve as the basis for the linking hypothesis. Only a small subset of A and C concepts will lead to new discoveries, and the sheer breadth of science means numerous false-positive links are likely to emerge. In fact, Swanson’s linking is best understood not as an objective literature-based discovery method, but as a method for generating post-hoc evidence to support subjective speculation (Kostoff et al., 2009). Although he tried to shroud his hypotheses in an objective algorithm, Swanson’s choices of A and C concepts were highly personal. Swanson himself had Raynaud’s syndrome and frequently suffered from migraine headaches (Smalheiser, 2017). And although he was an avid runner throughout his entire life, completing a half-marathon in 2004 at the age of 80, his chronic atrial fibrillation would in 2007 cause a series of strokes that ultimately ended both his running and scientific careers (Smalheiser, 2017). Just like every other scientist, Swanson came to the literature with his own ailments and observational priors, breaking our model of the purely literature-driven scientist.
Literature-based discovery within the life sciences
Despite the hidden personal factors influencing Swanson’s early success, the promise of pure literature-based discovery in conjunction with the growing availability of bibliographic databases like MEDLINE fueled the growth of literature-based discovery as a field from the 1990’s into the present. While the promise of literature-based discovery as an automator of the scientific process is technically agnostic to field, apart from a few outliers (Valdés-Pérez, 1999; Kostoff, Solka, Rushenberg, & Wyatt, 2008; Gordon, Lindsay, & Fan, 2002), the early history of literature-based discovery is dominated by life science applications. From Gordon & Dumais’s (1998) latent semantic indexing alternative attempt at replicating Swanson’s Raynaud’s insights, to the potential genetic association between Parkinson’s and Crohn’s disease uncovered through bibliographic coupling (Kostoff, 2014), literature-based discovery techniques have been consistently motivated and benchmarked by biomedical data (Frijters et al., 2010; Sebastian, Siew, & Orimaye, 2017).
This close relationship between literature-based discovery and the biomedical sciences is not just a historical coincidence. Over 37 million papers are assigned to the subject “Medicine” within the OpenAlex bibliographic database, the most of any subject. In the United States, the life sciences also receive the largest share of federal R&D dollars by a wide margin. Taken together, these two points indicate that the biomedical domain is quite expansive, increasing the likelihood of latent knowledge being buried in the literature.
The scientific process within the life sciences is also amenable to literature-based discovery techniques. Due to the enormous complexity and measurement obstructions inherent to biological systems, the life sciences are often consigned to measuring co-occurrence statistics of various observable factors from which they can begin to build mechanistic theory. For example, does the presence of a particular gene in a population lead to increased disease susceptibility within that population? Does the observance of a comorbidity indicate an underlying causal relationship of the comorbidity on a diagnosis? Is there evidence that a drug approved to treat a particular condition could be repurposed and re-sold to treat another? These types of co-occurrence observations are central to scientific progress in the life sciences, and literature-based discovery techniques automate the detection of these correlations.
Because of this close structural relationship between the co-occurrence mining performed by literature-based discovery methods and the correlational insights sought by the life sciences, by the end of the 2010’s nearly all literature-based discovery methods and validation examples were derived from the life sciences (Sebastian, Sew, & Orimaye, 2017). To support of these emerging automated discovery techniques, the life sciences are home to a growing collection of biocurated databases which extract and restructure information from the literature and additional experimental sources for usage in literature-based discovery systems. Databases like UniProt, which collects protein sequence and function information, or ChEMBL, a curated database of bioactive molecules with drug-like properties, have streamlined the extraction and curation of the literature with an eye towards particular research and development applications. In this way, the life sciences have built a system for rapidly positioning literature within an ontological structure that can be quickly queried for co-occurrence insights, though these insights now come from data which has been extracted and abstracted from the original academic text. This does cause one to wonder whether this explosion of biocurated ontologies has aided in autonomously generating scientific observations, or whether these ontologies have simply created a more abstract domain wherein latent knowledge now resides between ontological boundaries, and discovery opportunities are contingent on the promptness and correctness of the biocuration process. Given the impressive developments within natural language processing over the past decade, perhaps one could better peer across these ontological boundaries by returning to the scientific text itself?
An example in materials science
In 2019, Tshitoyan et al. (2019) showed that purely text-based literature discovery methods can be effective in the materials science literature. Using 3.3 million materials science abstracts published between 1922 and 2018, the authors computed vector representations of each of the approximately 500,000 words in the abstract corpus using the skip-gram with negative sampling variation of the Word2Vec algorithm (Mikolov, Sutskever, Chen, Corrado, & Dean, 2013). This produces a 200-dimensional vector representation of each word and, crucial to the results of the paper, each chemical formula found throughout the abstracts.
Although the Word2Vec embedding algorithm is completely self-supervised and endowed with no prior materials science information, the resulting organization of the embeddings reflect real physical properties. Not only do compounds with similar physical properties cluster together in this embedding space, but vector arithmetic in this space also takes on a physical meaning. For example, the authors find indications of “oxide of” and “structure of” dimensions in the space which physical facts like zirconium dioxide is the principal oxide of zirconium, or that “antiferromagnetic” is approximately the embedding that results from removing the ferromagnetic alloy “NiFe” from the vector for “ferromagnetic” and adding back “IrMn”, an antiferromagnetic alloy: “antiferromagnetic ~ ferromagnetic - NiFe + IrMn”.
Noting these vector-space indications of physical material properties, Tshitoyan et al. (2019) turned towards discovery, investigating the extent to which new materials with particular desirable functional properties can be discovered based on their similarity in the embedding space to vectorized representation of the property. Focusing on thermoelectric potential, the authors computed the cosine similarity of the vector representations of all material formulas to the word “thermoelectric”, thereby producing a ranking of materials based on their perceived similarity to the property based on the text corpus. Interestingly, the rank of these materials in their similarity to “thermoelectric” was found to correlate with the power factor of these materials, indicating not only a relationship to thermoelectric potential, but also a magnitude. While many of the most similar materials are well-known thermoelectrics, some of the top-ranked results have likely never had their thermoelectric potential reported in the literature, indicating prospective new thermoelectric materials.
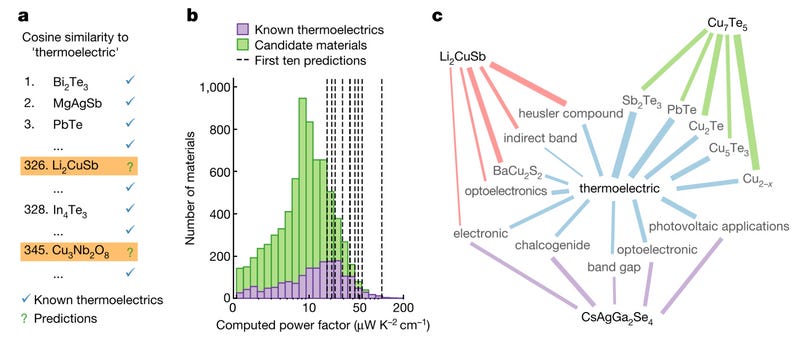
To validate this embedded literature-based discovery method’s capacity for discovering prospective thermoelectric materials, Tshitoyan et al. (2019) simulate the historical thermoelectric discovery process and measure the extent to which their embeddings anticipate the discovery of new thermoelectrics based on the ranked similarity of these material embeddings to the “thermoelectric” embedding in each year. By computing these word embeddings with the most recent publication cutoff year taken at each year between 2001 and 2018, the authors can “backtest” the performance of these embeddings as a basis for thermoelectric literature-based discovery. They find that the top-50 most similar materials to “thermoelectric” were on average 8x more likely than another randomly chosen material to go on to be studied as thermolectrics within the proceeding five years and 3x more likely than another randomly chosen material with a simulated non-zero bandgap (indicating thermoelectric potential). Three of the method’s top-five ranked prospective thermoelectric materials in 2009 would go on to have their thermoelectric properties positively measured in the literature within the proceeding decade.

Nothing new under the sun
The success of this embedded literature-based discovery method has given rise to a spate of follow-on literature-based discovery models since proposed for the materials science domain (Huang & Cole, 2022; Mai, Le, Chen, Winkler, & Caruso, 2022; Choudhary et al., 2022). While this excitement surrounding this machine learned, NLP-based techniques appears methodologically novel, the embedding-based approach to material function discovery described by Tshitoyan et al. (2019) is merely a modern spin on the prior keyword-based methods pioneered by Swanson and many others in the literature-based discovery community.
To see this, we require a bit of background theory. In 2014, Levy & Goldberg (2014) showed that the Word2Vec word embedding algorithm is actually an approximation of more traditional factorization-based approaches to vectorizing text. In particular, they showed that Word2Vec approximately factorizes a matrix which encodes the pointwise-mutual information between each pair of words in the corpus. In Tshitoyan et al. (2019), this corresponds to a (500,000 x 500,000)-dimensional matrix, each element of which records the probability of observing two words together within a small window of the corpus relative to the product of the words’ individual observation probabilities. Deriving word vectors from a factorization of the pointwise mutual information matrix implies that the inner product between two word vectors—the operation underlying cosine similarity—corresponds to an approximation of the increase or decrease in statistical co-occurrence observed between word pairs within the corpus relative to their expected rate of co-occurrence if they were merely independent.
Recall that in 1998, Gordon & Dumais (1998) proposed latent semantic indexing as a potential literature-based discovery method. Latent semantic indexing, too, is a factorization method for deriving vector embeddings. In particular, it computes a low-rank representation of each word by factorizing the word-document matrix of a corpus. This means the inner product between two words approximates their correlation within documents throughout the corpus. While this correlation is not exactly equivalent to the pointwise mutual information captured by the Word2Vec algorithm, both represent a measure of pairwise relatedness between words (covariance in correlation, joint probability in pointwise mutual information) relative to the product of their individual statistics (standard deviation in correlation, marginal probability in pointwise mutual information). So although the Word2Vec approach to literature-based discovery in Tshitoyan et al. (2019) provides additional representational power through its nonlinear approximation of pointwise mutual information, the statistical motivation is very similar to the latent semantic indexing approach proposed over twenty years earlier. Moreover, Swanson’s original ABC procedure may be understood as nothing more than a procedural search across keywords in the literature based on their correlation between document corpuses. What’s old is new again.
In noting the similarity between modern literature-based discovery methods which use sophisticated machine learning representations and the traditional correlation-derived methods, I do not mean to imply that these modern iterations lack novelty. The robustness and expressivity of these newer representation learning models makes them much more capable in detecting nuanced latent relationships within the literature in comparison to their predecessors. However, when provided only a corpus of scientific text, our statistical insight is fundamentally constrained to the measurement of observed joint relationships relative to their expected prevalence under some null model of the distribution of scientific text. This constraint is universal, likely holding even with modern large language models (Huh, Cheung, Wang, & Isola, 2024), so although modern NLP techniques can incrementally improve the existing literature-based discovery paradigm, a major breakthrough in this area may require an adventure beyond the existing statistical co-occurrence framework.
Just ask the LLM
The frontier of literature-based discovery research is becoming increasingly dominated by techniques which rely on large language models (LLMs) to generate new discoveries (Qi et al., 2023; O’Brien et al., 2024; Baek, Jauhar, Cucerzan, & Hwang, 2024; Lu et al., 2024). This progression is sensible because, broadly construed, the largest LLMs with billions of tunable parameters may be interpreted as a functional compression of all online human knowledge. In this way, LLMs are our closest technological approximation to the scientist-as-reader model discussed above: extremely well-read but somewhat lacking in observational capacity and a robust understanding of the physical world.
Because they can query and report scientific knowledge through natural language, LLMs make an especially exciting—and dangerous—literature-based discovery model. Natural language is the primary medium of science, as the “Report conclusions” step of the scientific method is typically facilitated by the publication of a paper. By aligning literature-based discovery techniques closer to the default medium of the scientific process, one hopes that these discoveries could be located and disseminated more efficiently. While the new thermoelectric materials unearthed by Tshitoyan et al. (2019) were derived from scientific text, the Word2Vec model only produces a latent vector representation of the materials and words strewn throughout the academic corpus. Thus, it was on the authors of the paper to construct a metahypothesis (there are undiscovered thermoelectrics that can be derived from the literature) and a methodology for evaluating potential sub-hypotheses (computing and ranking cosine similarity of materials with “thermoelectric”) in order to produce a literature-based discovery pipeline. The text-native nature of the LLM, an encoder which interprets textual prompts paired with a decoder which generates text, promises to streamline this text-based discovery process by circumventing the intermediate calculations in the latent space. Just ask the LLM where the new thermoelectrics are hiding.
The use of LLMs for literature-based discovery comes with its own set of concerns. By removing intermediate meta-hypothesizing steps like choosing A and C concepts or constructing a thermoelectric potential ranking, LLM-based methods place a larger burden on humans to validate the discoveries proposed by the LLMs, as the output of these models are typically unconstrained and may contain nuanced errors. LLMs are also traditionally trained on an autoregressive task and therefore learn representations which help to generate high-probability outputs. Novel scientific insights, by definition, occupy a low-probability region of semantic space, so encouraging LLMs to explore these low-density regions while remaining cogent is an open and fundamental problem for literature-based discovery which rely on LLMs to generate novelty. Even if an optimal tradeoff between LLM exploration and cogency were to be found, LLMs are still subject to the same correlational inferential constraints of all of the literature-based discovery methods which came before them (Huh, Cheung, Wang, & Isola 2024).
So, how much new knowledge is hidden in scientific text?
This is a difficult question to answer from the literature alone, and one that may not have an answer. Although Swanson himself was able to “discover” a number of interesting connections between seemingly disparate topics of study, we know that his health provided him implicit prior knowledge for how to filter through correlated literature. The prior knowledge bias required to pick out the link between Raynaud’s and fish oil purely from co-occurrence statistics has been noted by many different literature-based discovery techniques during attempts to replicate Swanson’s original discovery (Gordon & Lindsay, 1996; Kostoff et al., 2009). Due to a general lack (or even impossibility) of datasets which can be used to benchmark these literature-based discovery techniques (Sebastian, Siew, & Orimaye, 2017), it is generally difficult to know which literature-derived discoveries are a result of pure statistical insight, and which are tainted by the authors’ prior knowledge.
Even if we take these prior discoveries for granted, we still don’t know how much knowledge is hidden in the present literature. Probabilistically, there are a multitude of combinations of topics or words which one could find in a snapshot of today’s scientific literature which, according to any of the literature-based discovery methods discussed above, would imply hidden scientific potential. However, validating the extent to which this potential corresponds to novel and meaningful knowledge requires experimental investigation, most likely by human hands.
And even if there were a particularly effective literature-based discovery method which could locate latent scientific knowledge within the literature, it is unlikely that we would see its public publication. The reward structures of science and technology are such that, if one had an oracle which could indicate high-potential scientific insights, a rational actor would derive much higher reward by publishing or patenting each insight, as opposed to publishing details about the inner workings of the oracle. In this way, the literature-based discovery literature has a similar risk of publication suppression that currently plagues the quantitative finance literature. In quantitative finance, successful market trading strategies are rarely published due to immense opportunity cost introduced by the alternative to publication: operationalizing this trading strategy and profiting from its novelty directly. Because new discoveries in the life sciences—like emerging diagnostic markers for diseases or new candidate populations for drug repurposing—are even more lucrative than a profitable trading strategy, the adoption of literature-based discovery techniques in the life sciences means that, even if a particular technique works well in this area, we are unlikely to see its publication until its ability to highlight potential discoveries deteriorates below the expected value of publishing the model itself.
I will continue to update this article as the literature-based discovery literature evolves, but with each update one must ask two questions: one, what prior knowledge has been used to choose these particular discoveries from the much larger space of possible hypotheses presented by the model, and two, if this literature-based discovery technique works well then why is it being publishing instead of the discoveries themselves?
References
Baek, J., Jauhar, S. K., Cucerzan, S., & Hwang, S. J. (2024). Researchagent: Iterative research idea generation over scientific literature with large language models. arXiv Preprint arXiv:2404.07738.
Choudhary, K., DeCost, B., Chen, C., Jain, A., Tavazza, F., Cohn, R., … others. (2022). Recent advances and applications of deep learning methods in materials science. Npj Computational Materials, 8(1), 59.
DiGiacomo, R. A., Kremer, J. M., & Shah, D. M. (1989). Fish-oil dietary supplementation in patients with raynaud’s phenomenon: A double-blind, controlled, prospective study. The American Journal of Medicine, 86(2), 158–164.
Frijters, R., Van Vugt, M., Smeets, R., Van Schaik, R., De Vlieg, J., & Alkema, W. (2010). Literature mining for the discovery of hidden connections between drugs, genes and diseases. PLoS Computational Biology, 6(9), e1000943.
Gallai, V., Sarchielli, P., Coata, G., Firenze, C., Morucci, P., & Abbritti, G. (1992). Serum and salivary magnesium levels in migraine. Results in a group of juvenile patients. Headache: The Journal of Head and Face Pain, 32(3), 132–135.
Gordon, M. D., & Dumais, S. (1998). Using latent semantic indexing for literature based discovery. Journal of the American Society for Information Science, 49(8), 674–685.
Gordon, M. D., & Lindsay, R. K. (1996). Toward discovery support systems: A replication, re-examination, and extension of swanson’s work on literature-based discovery of a connection between raynaud’s and fish oil. Journal of the American Society for Information Science, 47(2), 116–128.
Gordon, M., Lindsay, R. K., & Fan, W. (2002). Literature-based discovery on the World Wide Web. ACM Transactions on Internet Technology (TOIT), 2(4), 261–275.
Huang, S., & Cole, J. M. (2022). BatteryBERT: A pretrained language model for battery database enhancement. Journal of Chemical Information and Modeling, 62(24), 6365–6377.
Hubbard, K. E., & Dunbar, S. D. (2017). Perceptions of scientific research literature and strategies for reading papers depend on academic career stage. PloS One, 12(12), e0189753.
Huh, M., Cheung, B., Wang, T., & Isola, P. (2024). Position: The platonic representation hypothesis. Forty-First International Conference on Machine Learning.
Huh, M., Cheung, B., Wang, T., & Isola, P. (2024). Position: The platonic representation hypothesis. Forty-First International Conference on Machine Learning.
Kostoff, R. N. (2014). Literature-related discovery: Common factors for parkinson’s disease and crohn’s disease. Scientometrics, 100(3), 623–657.
Kostoff, R. N., Block, J. A., Solka, J. L., Briggs, M. B., Rushenberg, R. L., Stump, J. A., … Wyatt, J. R. (2009). Literature-related discovery. Annual Review of Information Science and Technology, 43(1), 1–71.
Kostoff, R. N., Solka, J. L., Rushenberg, R. L., & Wyatt, J. A. (2008). Literature-related discovery (LRD): Water purification. Technological Forecasting and Social Change, 75(2), 256–275.
Levy, O., & Goldberg, Y. (2014). Neural word embedding as implicit matrix factorization. Advances in Neural Information Processing Systems, 27.
Lu, C., Lu, C., Lange, R. T., Foerster, J., Clune, J., & Ha, D. (2024). The AI Scientist: Towards Fully Automated Open-Ended Scientific Discovery. Retrieved from https://arxiv.org/abs/2408.06292
Mai, H., Le, T. C., Chen, D., Winkler, D. A., & Caruso, R. A. (2022). Machine learning for electrocatalyst and photocatalyst design and discovery. Chemical Reviews, 122(16), 13478–13515.
Mikolov, T., Sutskever, I., Chen, K., Corrado, G. S., & Dean, J. (2013). Distributed representations of words and phrases and their compositionality. Advances in Neural Information Processing Systems, 26.
Mont, L., Elosua, R., & Brugada, J. (2009). Endurance sport practice as a risk factor for atrial fibrillation and atrial flutter. Europace, 11(1), 11–17.
O’Brien, T., Stremmel, J., Pio-Lopez, L., McMillen, P., Rasmussen-Ivey, C., & Levin, M. (2024). Machine Learning for Hypothesis Generation in Biology and Medicine: Exploring the latent space of neuroscience and developmental bioelectricity. Digital Discovery, 3(2), 249–263.
Qi, B., Zhang, K., Li, H., Tian, K., Zeng, S., Chen, Z.-R., & Zhou, B. (2023). Large Language Models are Zero Shot Hypothesis Proposers. NeurIPS 2023 Workshop on Instruction Tuning and Instruction Following. Retrieved from https://openreview.net/forum?id=EAuteBjTMw
Sebastian, Y., Siew, E.-G., & Orimaye, S. O. (2017). Emerging approaches in literature-based discovery: Techniques and performance review. The Knowledge Engineering Review, 32, e12.
Smalheiser, N. R. (2017). Rediscovering don swanson: The past, present and future of literature-based discovery. Journal of Data and Information Science, 2(4), 43–64.
Smalheiser, N. R., Torvik, V. I., Bischoff-Grethe, A., Burhans, L. B., Gabriel, M., Homayouni, R., … others. (2006). Collaborative development of the arrowsmith two node search interface designed for laboratory investigators. Journal of Biomedical Discovery and Collaboration, 1, 1–18.
Swanson, D. R. (1986). Fish oil, raynaud’s syndrome, and undiscovered public knowledge. Perspectives in Biology and Medicine, 30(1), 7–18.
Swanson, D. R. (1986). Undiscovered public knowledge. The Library Quarterly, 56(2), 103–118.
Swanson, D. R. (1987). Two medical literatures that are logically but not bibliographically connected. Journal of the American Society for Information Science, 38(4), 228–233.
Swanson, D. R. (1988). Migraine and magnesium: Eleven neglected connections. Perspectives in Biology and Medicine, 31(4), 526–557.
Swanson, D. R. (1989). A second example of mutually isolated medical literatures related by implicit, unnoticed connections. Journal of the American Society for Information Science, 40(6), 432.
Swanson, D. R. (1990). Somatomedin c and arginine: Implicit connections between mutually isolated literatures. Perspectives in Biology and Medicine, 33(2), 157–186.
Swanson, D. R. (2006). Atrial fibrillation in athletes: Implicit literature-based connections suggest that overtraining and subsequent inflammation may be a contributory mechanism. Medical Hypotheses, 66(6), 1085–1092.
Swanson, D. R., & Smalheiser, N. R. (1997). An interactive system for finding complementary literatures: A stimulus to scientific discovery. Artificial Intelligence, 91(2), 183–203.
Tshitoyan, V., Dagdelen, J., Weston, L., Dunn, A., Rong, Z., Kononova, O., … Jain, A. (2019). Unsupervised word embeddings capture latent knowledge from materials science literature. Nature, 571(7763), 95–98.
Uzzi, B., Mukherjee, S., Stringer, M., & Jones, B. (2013). Atypical combinations and scientific impact. Science, 342(6157), 468–472.
Valdés-Pérez, R. E. (1999). Principles of human—computer collaboration for knowledge discovery in science. Artificial Intelligence, 107(2), 335–346.
Weitzman, M. L. (1998). Recombinant growth. The Quarterly Journal of Economics, 113(2), 331–360.



